Hi! I'm Binghao, a research fellow in Artificial Intelligence (AI) for healthcare at UCL Cancer Institute.
About Me

I am Binghao Chai (柴 秉浩), a Research Fellow in Artificial Intelligence (AI) for Healthcare at UCL Cancer Institute (Flanagan Lab). My research focuses on the development and application of AI and computer vision techniques for digital pathology and cancer diagnostics. Prior to this, I was a Postdoctoral Computer Vision Scientist at Queen Mary University of London (QMUL), working at Draviam Lab under the UKRI Innovate UK Knowledge Transfer Partnership (KTP) scheme in collaboration with ZEISS. I earned my PhD in Computer Science and Digital Pathology from University College London (UCL), supervised by Dr Kevin Bryson and Prof Nischalan Pillay. I also holds an MSc Software Engineering (UCL) and a BSc Computer Science (the University of Manchester).
My research interests are applied-AI/ML, biomedical imaging, computational pathology and cell biology imaging analysis. I take an entrepreneurial approach to my career and all single work, and I have driven change and innovation in every role by focusing on key goals and applying a growth mindset.
Research
AI for soft tissue sarcoma pathology
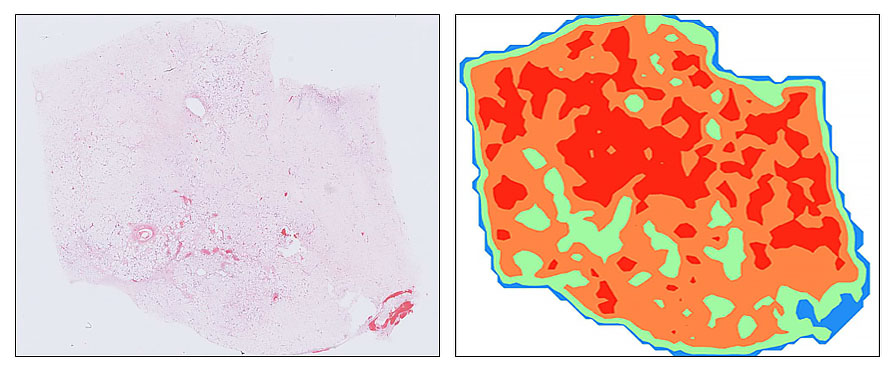
My research focuses on applying advanced AI techniques to the diagnosis and subtyping of soft tissue sarcomas and their mimics, with a particular emphasis on computational classification from digitised histopathology. Soft tissue sarcomas are rare, morphologically diverse tumours that present significant diagnostic challenges, often requiring expert review and ancillary testing. By leveraging modern deep learning techniques, including state-of-the-art foundation models, I aim to improve diagnostic accuracy, efficiency, and generalisability in real-world clinical settings.
One strand of my work, conducted in collaboration with Google Health and The Alan Turing Institute, explores how variations in tissue staining and scanning protocols across laboratories affect the performance of AI models in pathology. Using sarcoma pathology as a challenging test case, this project evaluates multiple pathology foundation models to understand their robustness and adaptability. The goal is to provide insights that can inform the development of AI systems capable of consistent and reliable performance across diverse clinical environments (available here).
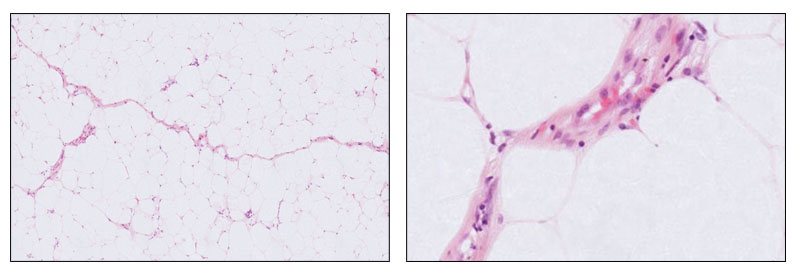
Alongside this, I also investigate tumour classification approaches in soft tissue pathology, including methods that address common diagnostic challenges. An example is an earlier work (2019–2022, available here) on developing computational pipelines to distinguish between benign lipomas and malignant atypical lipomatous tumours from whole slide images, a task that often requires detailed morphological assessment and specialist review. These complementary research directions, exploring colour-related techniques as well as advancing tumour classification methods, aim to pave the way for more reliable, adaptable, and clinically useful AI tools in digital pathology.
Microscopy time-lapse image analysis of dynamic cellular structures
In addition to my work in digital pathology, I also conduct research in the analysis of high-resolution microscopy time-lapse images, particularly for studying the dynamic subcellular structures of live-cells. Advances in live-cell imaging technologies now allow researchers to capture large volumes of 3D and time-lapse data, revealing intricate biological processes in unprecedented detail. However, the complexity of these datasets (with structures moving in three dimensions, changing shape over time, and often existing in crowded environments and more) makes automated analysis both computationally challenging and essential for high-throughput studies. My work in this area aims to design and implement computational frameworks that enable robust, scalable, and accurate tracking of such structures, helping to unlock new biological insights while reducing the reliance on labor-intensive manual annotation.
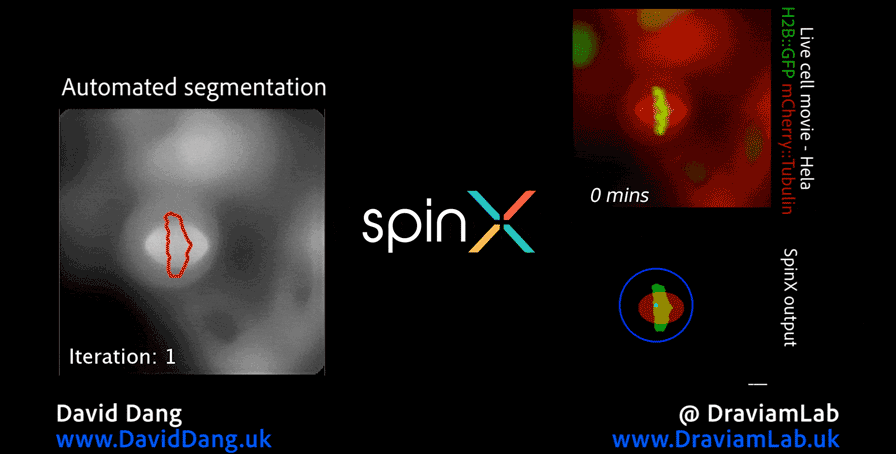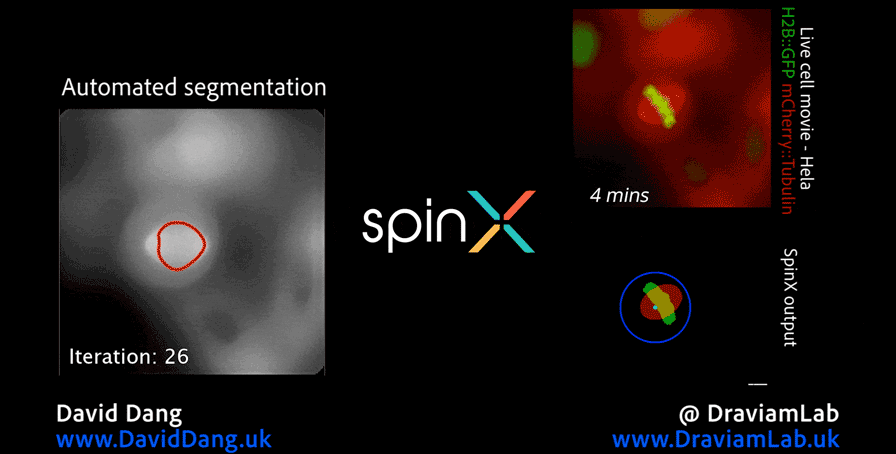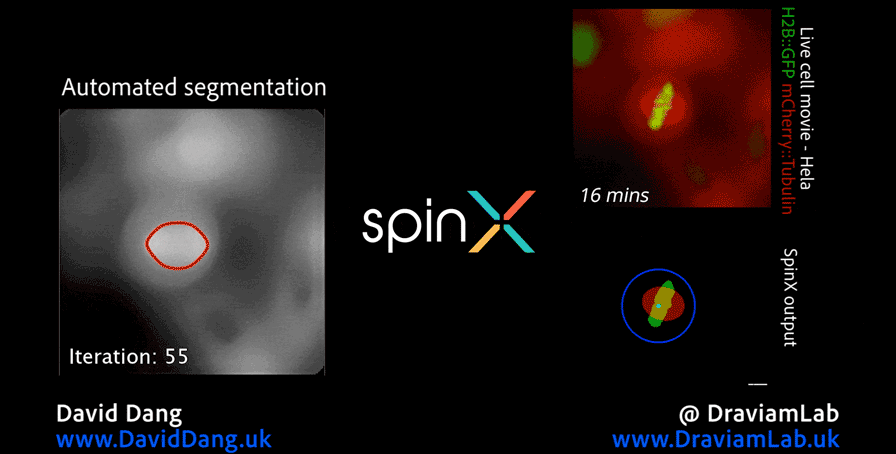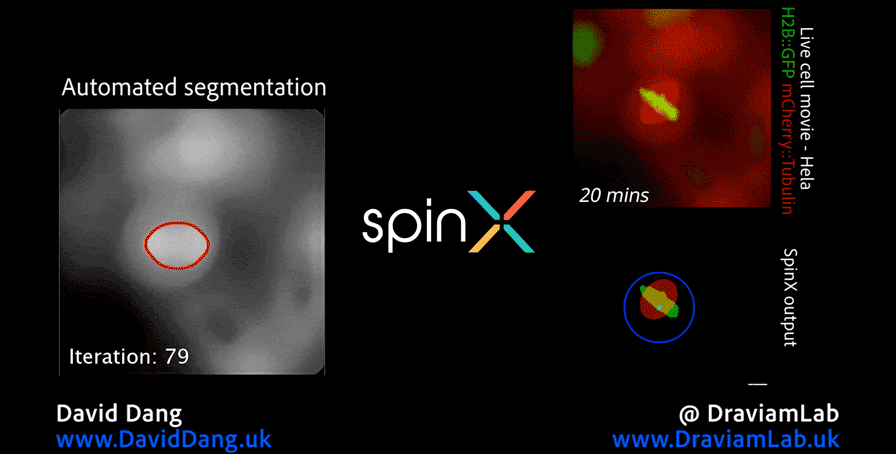
As part of this research, I led the development of Multi-SpinX (available here), an advanced computational framework for automated tracking of mitotic spindles and kinetochores in multicellular environments. During mitosis, the mitotic spindle (a dynamic microtubule-based structure) orchestrates the segregation of chromosomes, which are attached via the kinetochore at their centromeric regions. Both structures undergo complex and often independent movements in 3D space over time, making them particularly challenging to track in crowded or high-throughput imaging experiments. Multi-SpinX extends the capabilities of the original SpinX system (available here), which was limited to single-cell metaphase analysis. The framework was developed in collaboration with Prof. Viji Draviam (Queen Mary University of London), Prof. Kozo Tanaka (Tohoku University), ZEISS, and other colleagues. Multi-SpinX is now integrated into ZEISS arivis Pro and ZEISS arivis Cloud, and provides a scalable solution for researchers studying spindle–kinetochore dynamics, enabling richer quantitative analyses of mitosis in complex multicellular contexts. Other related publications are available here (review paper, Trends in Cell Biology) and here (interview article, infocus Magazine from Royal Microscopical Society).
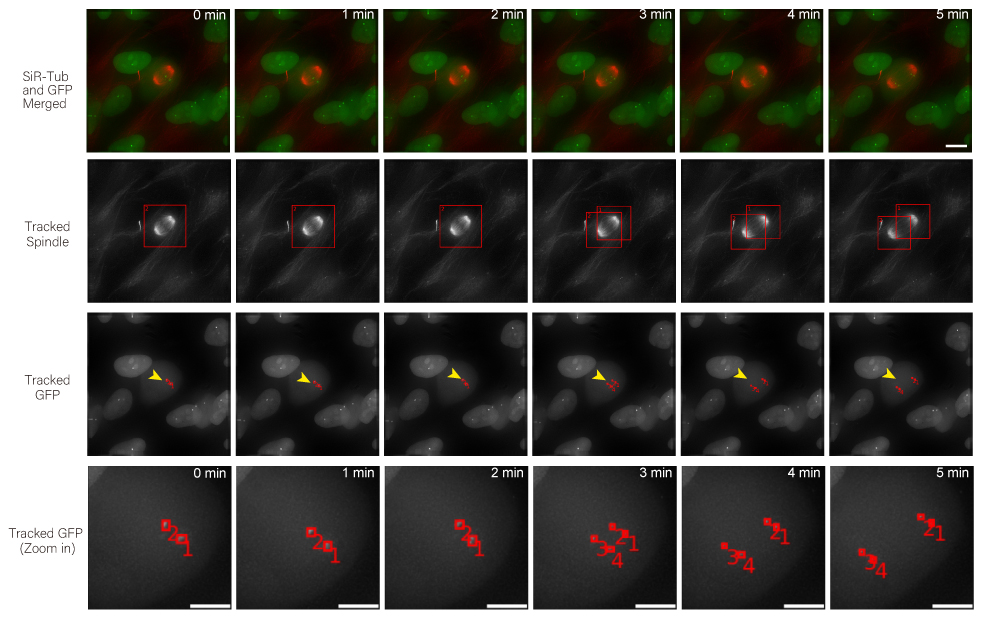
Time-lapse images of consecutive frames illustrate the complex process of kinetochore tracking overlaid with spindle tracking. The frames are organised as follows: the original movie as a merge of the spindle (red) and kinetochore marker (centromere marker for chromosome 16, in green) displays the representative consecutive frames (first row), the tracked spindle with bounding boxes illustrates the spindle tracking outcome (second row), the tracked GFP with bounding boxes (red, marked by yellow arrow) depicts the kinetochores being tracked, with bounding boxes delineating the track location (third row), and the magnified consecutive frames highlighted with yellow arrows in the third row showcase sister kinetochores with two particle IDs (1 and 2) segregated into four kinetochores (1,2,3 and 4). Scale bar: 10 μm.
Teaching and Supervision
Research Project Supervision
Research project supervisor to Muntaqa Shahana Choudhury (PhD candidate at QMUL), Saakshi Sanjay Jain and Sana Zakir Piracha (both undergrad students at QMUL) for the Multi-SpinX project (2024).
Master's and Bachelor's Thesis Supervision
Alexia-Cristina Maharea, bachelor's thesis at QMUL (2024). Thesis: Assessing multi-spindle tracker to improve and support development.
Alan Jeyaram Sounthararajah, bachelor's thesis at QMUL (2024). Thesis: Improving SpinX: the multispindle tracker.
Jeel Maheshkumar Prajapati, master's thesis at QMUL (2023). Thesis: Artificial Intelligence tools to advance drug discovery screens: developing a nuclear atypia tracker using a deep learning framework at Zeiss arivis Cloud.
Teaching Assistant
Requirement Engineering and Software Architecture (COMP0101) at UCL (2021/22, 2020/21, 2019/20, 2018/19).
Software Abstraction and Systems Integration (COMP0102) at UCL (2021/22, 2020/21, 2019/20, 2018/19).
Validation and Verification (COMP0103) at UCL (2020/21).
Systems Engineering (COMP0016) at UCL (2020/21).
Machine Learning for Domain Specialists (COMP0142) at UCL (2019/20).
Software Engineering (COMP0071) at UCL (2018/19).
Miscellaneous
Binghao and his classic music
Gallery
Old News
Nov. 2025, I gave a talk From Fading stains to foundation models: adapting digital pathology to real world complexity at the National Institute of Theory and Mathematics in Biology (NITMB, Chicago).
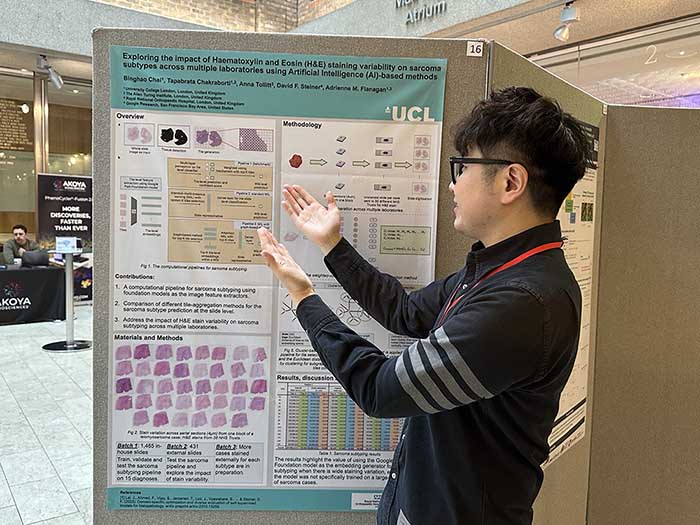
Jan. 2025, I presented Exploring the impact of Haematoxylin and Eosin staining variability on sarcoma subtypes across multiple laboratories using Artificial Intelligence (AI)-based methods (poster) at Pathological Society 2025 Joint Winter Meeting with The Royal Society of Medicine.
Apr. 2024, I joined Flanagan Lab at UCL Cancer Institute as a research fellow in AI for healthcare.
Aug. 2023 to Jul. 2024, I co-hosted a series of "International Symposium on Genomic Instability and Impact of Genetic Variants" workshops and presented "AI in biomedical image and movie analysis" and "Multi-SpinX: An Advanced Framework for Automated Tracking of Mitotic Spindles and Kinetochores in Multicellular Environments" talks and posters at Kanagawa Institute of Technology (Tokyo Japan, Aug. 2023), China Agricultural University (Beijing China, Sept. 2023), Queen Mary University of London (London UK, Dec. 2023), Queen Mary University of London (London UK, Mar. 2024), and Queen Mary University of London (London UK, Jul. 2024)
Mar. 2023, I passed my final viva and was awarded a PhD degree.
Feb. 2023, I joined Draviam Lab at Queen Mary University of London as a postdoctoral computer vision scientist.
Jul. 2019, I passed my upgrade viva and became a PhD candidate.
Apr. 2019, I presented "An emerging role for machine learning in the classification of tumours of fat: distinguishing benign from malignant" at the 15th European Congress on Digital Pathology (ECDP 2019).
Jan. 2018, I joined Bryson Lab in collobration with Pillay Lab at UCL as a PhD student.